After learning that the heritability of traits tend to be quite high, many act as if family members should be highly similar, physically and behaviorally. But this is not always the case.
Under conditions of no assortative mating (meaning the phenotypes of mates don’t correlate), mates are as similar to each other as strangers, and the genetic variance among their offspring is half that of the general population.
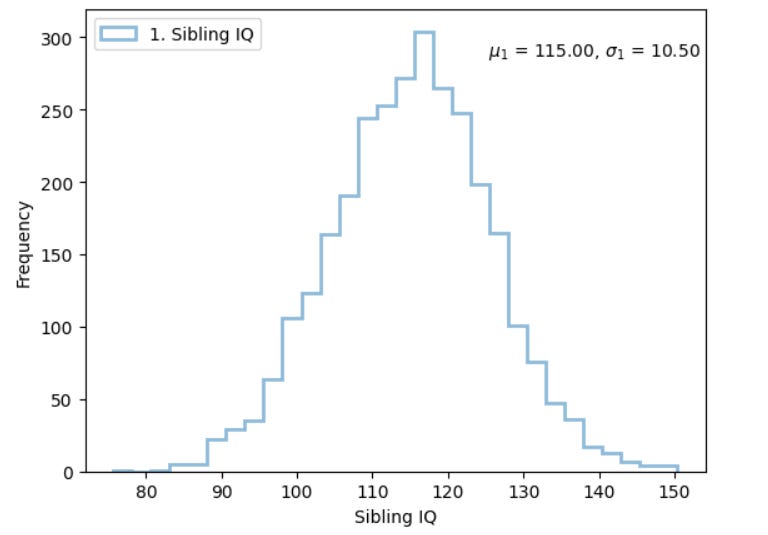
This means, for a trait that is 100% heritable, the standard deviation of that trait is SQRT(0.50) =~ 70% of that of the general population. This is, for example, 15 * 0.70 = 10.5 IQ points.
If the genetic parental midpoint were 115, the distribution of sibling IQ gene scores would look like this:
As you can see, under conditions of no assortative mating, it is relatively common for one sibling to be genetically 95 IQ and one sibling to be genetically 135 IQ, even if the parents are midwits.
Short Term Assortative Mating Doesn’t Change This
SIZE = 2000
ps = np.random.normal(.5,.15,SIZE)
ps[ps > 1] = .999
ps[ps < 0] = 0.01
fifties = [0.5 for i in range(SIZE)]
NUM_FAMS = 500
vars = []
parents = np.zeros((NUM_FAMS*2, SIZE))
for _ in range(NUM_FAMS):
print(_/NUM_FAMS, end="\r")
NUM_KIDS = 500
m1 = np.random.binomial(1, ps)
m2 = np.random.binomial(1, ps)
f1 = np.random.binomial(1, ps)
f2 = np.random.binomial(1, ps)
parents[2*_] = m1+m2
parents[(2*_)+1] = f1+f2
kids = np.zeros((NUM_KIDS, SIZE)) #row: kid genome. col: genes
for i in range(NUM_KIDS):
samples = np.random.binomial(1, fifties)
samples2 = np.random.binomial(1, fifties)
kid1 = np.where(samples == 0, m1, m2) # same as kid1 = np.array([m1[idx] for idx in range(SIZE) if (samples[idx] == 0) else (m2[idx]) ]) but written in C
kid2 = np.where(samples2 == 0, m1, m2) #make m1,m2 for perfect assortative mating, f1,f2 for random mating
kids[i] = kid1 + kid2
varst = np.var(kids, axis=0)
vars.append( np.sum(varst))
vars = np.array(vars)
parentvarl = np.var(parents, axis=0)
parentvar = np.sum(parentvarl)
kidvar = vars.mean()
print(parentvar, kidvar, kidvar/parentvar)
#output: 910.1713849999999 454.36151949600014 0.4992043553379787I intuited that short term assortative mating would produce more similar siblings, but math and simulation contradict this. Above is code showing that if a large sibling batch is produced, having genetically identical mothers and fathers, the variance of sibling gene scores is still half that of the general parental population.
The math I came up with goes like this: consider a gene locus. Each sibling will have two copies at that locus: one from their mother, and one from their father. But pretend for a moment you know which locus is from the mother and which is from the father for each sibling. The total variance at this half-locus in the parental population is pq, where p is the probability of having a 1 at that locus and q is the probability of having a 0.
Among siblings over all families, the variance of this locus is 0 for families who have a homozygous parents for that locus, and 1/4 for those who have a heterozygous parent for that locus, because have the siblings have 1 and half have 0, and 0.5*0.5 = 1/4.
So
Now sum over all loci and you get V(G|S) = 1/2 V(G). In this model it does not matter whether or not the other parent has the same genes as the first one; the variance among siblings at every locus will always be 1/2 of that of the parent generation’s population variance, assuming the parents are at Hardy-Weinberg equlibrium1.
Apparently, over a number of generations, assortative mating leads to more homozygosity which leads to there being a lower proportion of variance among family members. However, the effects appear quite small for most traits.
Conclusion
The first chart here is a good approximation of the the spread of full sibling gene scores. In terms of IQ, full siblings can easily be more than 15 points apart genetically, and kids can easily be 10 or 20 IQ points higher or lower than their parents genetically.
As a commenter points out below, assortative mating will increase the overall variance for the kid generation. But a batch of siblings will always have 1/2 the additive variance of the parent generation. So if kid generation’s variance > parent generation’s variance, and sibling variance = 1/2 parent generation’s variance, then sibling variance < 1/2 kid generation’s variance.





I've gotten this question a number of times in fact.
You did your simulation wrong - it’s easy to see that under random mating that variance between midparents is half pop var & that within family variance provides the other half.
But if you rank order couples midparent variance is equal to population variance, assuming parent pedigrees are no similar & within family variance is unchanged, now a within family variance is now a third of population variance.
Basically you are doing your calculation relative to the wrong generation.